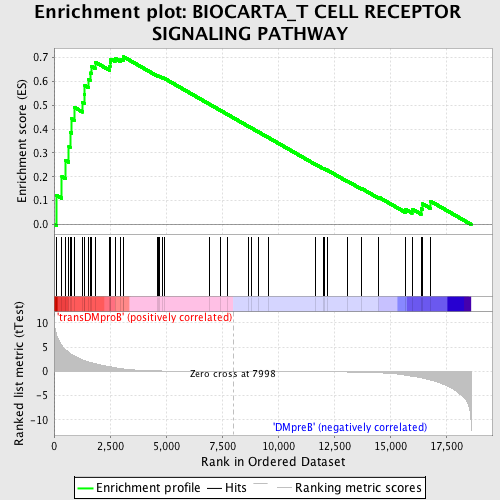
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | BIOCARTA_T CELL RECEPTOR SIGNALING PATHWAY |
| Enrichment Score (ES) | 0.70291126 |
| Normalized Enrichment Score (NES) | 1.6097862 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.17764363 |
| FWER p-Value | 0.702 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LAT | 17643 | 116 | 7.434 | 0.1217 | Yes | ||
| 2 | VAV1 | 23173 | 347 | 5.300 | 0.2005 | Yes | ||
| 3 | RALBP1 | 9725 | 507 | 4.482 | 0.2690 | Yes | ||
| 4 | PTPN7 | 11500 | 659 | 3.836 | 0.3269 | Yes | ||
| 5 | ARHGAP4 | 9343 | 716 | 3.651 | 0.3867 | Yes | ||
| 6 | LCK | 15746 | 786 | 3.491 | 0.4431 | Yes | ||
| 7 | ARFGAP3 | 22183 | 909 | 3.160 | 0.4909 | Yes | ||
| 8 | PPP3CC | 21763 | 1270 | 2.374 | 0.5124 | Yes | ||
| 9 | MAP3K1 | 21348 | 1355 | 2.218 | 0.5460 | Yes | ||
| 10 | PPP3CB | 5285 | 1370 | 2.196 | 0.5830 | Yes | ||
| 11 | GRB2 | 20149 | 1534 | 1.943 | 0.6077 | Yes | ||
| 12 | RELA | 23783 | 1612 | 1.845 | 0.6353 | Yes | ||
| 13 | JUN | 15832 | 1670 | 1.759 | 0.6625 | Yes | ||
| 14 | CAMK2B | 20536 | 1834 | 1.586 | 0.6810 | Yes | ||
| 15 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 2476 | 0.992 | 0.6636 | Yes | ||
| 16 | RAC1 | 16302 | 2500 | 0.969 | 0.6790 | Yes | ||
| 17 | MAPK3 | 6458 11170 | 2529 | 0.941 | 0.6937 | Yes | ||
| 18 | FOS | 21202 | 2723 | 0.766 | 0.6965 | Yes | ||
| 19 | RAF1 | 17035 | 2978 | 0.564 | 0.6925 | Yes | ||
| 20 | CD3D | 19473 | 3103 | 0.500 | 0.6944 | Yes | ||
| 21 | MAP2K1 | 19082 | 3106 | 0.498 | 0.7029 | Yes | ||
| 22 | ARHGAP6 | 24207 | 4626 | 0.093 | 0.6227 | No | ||
| 23 | ZAP70 | 14271 4042 | 4655 | 0.091 | 0.6228 | No | ||
| 24 | SOS1 | 5476 | 4708 | 0.086 | 0.6215 | No | ||
| 25 | CD3G | 19139 | 4851 | 0.076 | 0.6151 | No | ||
| 26 | PIK3R1 | 3170 | 4927 | 0.072 | 0.6123 | No | ||
| 27 | PTPRC | 5327 9662 | 6953 | 0.015 | 0.5035 | No | ||
| 28 | PLCG1 | 14753 | 7414 | 0.008 | 0.4789 | No | ||
| 29 | PIK3CA | 9562 | 7760 | 0.003 | 0.4604 | No | ||
| 30 | CD247 | 4498 8715 | 8697 | -0.009 | 0.4101 | No | ||
| 31 | CD3E | 8714 | 8819 | -0.011 | 0.4038 | No | ||
| 32 | SHC1 | 9813 9812 5430 | 9137 | -0.016 | 0.3870 | No | ||
| 33 | FYN | 3375 3395 20052 | 9588 | -0.022 | 0.3632 | No | ||
| 34 | PRKCA | 20174 | 11681 | -0.062 | 0.2516 | No | ||
| 35 | CD4 | 16999 | 12030 | -0.073 | 0.2341 | No | ||
| 36 | ARHGAP5 | 4412 8625 | 12065 | -0.074 | 0.2336 | No | ||
| 37 | TRB@ | 1157 | 12184 | -0.079 | 0.2286 | No | ||
| 38 | NCF2 | 14098 5151 9446 | 13091 | -0.124 | 0.1819 | No | ||
| 39 | ARHGAP1 | 6001 10448 | 13740 | -0.179 | 0.1501 | No | ||
| 40 | ARFGAP1 | 14705 2854 | 14482 | -0.279 | 0.1150 | No | ||
| 41 | MAP2K4 | 20405 | 15684 | -0.770 | 0.0636 | No | ||
| 42 | PPP3CA | 1863 5284 | 15979 | -0.995 | 0.0649 | No | ||
| 43 | NFKBIA | 21065 | 16384 | -1.316 | 0.0658 | No | ||
| 44 | MAPK8 | 6459 | 16452 | -1.403 | 0.0863 | No | ||
| 45 | HRAS | 4868 | 16802 | -1.753 | 0.0977 | No |

